Evaluation of probe sets
! This tutorial will be updated the next days !
This tutorial shows how to to evaluate a probeset with the Spapros evaluation pipeline. We examplary evaluate the probeset of 20 genes that was selected in our basic selection tutoral.
Import packages and setup
To run the notebook locally, create a conda environment using this yaml file:
conda create -f environment.yaml
[4]:
import scanpy as sc
import spapros as sp
[5]:
sc.settings.verbosity = 1
sc.logging.print_header()
print(f"spapros=={sp.__version__}")
scanpy==1.8.2 anndata==0.8.0 umap==0.5.2 numpy==1.21.5 scipy==1.7.3 pandas==1.3.5 scikit-learn==1.0.2 statsmodels==0.13.2 python-igraph==0.9.9 pynndescent==0.5.6
spapros==0.1.0
Load dataset
[6]:
adata = sc.datasets.pbmc3k()
adata_tmp = sc.datasets.pbmc3k_processed()
# Get infos from the processed dataset
adata = adata[adata_tmp.obs_names, adata_tmp.var_names]
adata.obs['celltype'] = adata_tmp.obs['louvain']
adata.obsm['X_umap'] = adata_tmp.obsm['X_umap']
del adata_tmp
# Preprocess counts and get highly variable genes
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, flavor="cell_ranger", n_top_genes=1000)
adata
[6]:
AnnData object with n_obs × n_vars = 2638 × 1838
obs: 'celltype'
var: 'gene_ids', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'log1p', 'hvg'
obsm: 'X_umap'
Get probesets
[7]:
#probeset = ['PPBP', 'HLA-DQA1', 'FCGR3A', 'GZMB', 'CD79A', 'NKG7', 'S100A8', 'CD2',
# 'HLA-DPB1', 'GZMK', 'LST1', 'TYROBP', 'MS4A1', 'GPX1', 'CCL5',
# 'HLA-DRB1', 'GNLY', 'FCN1', 'SDPR', 'AIF1', 'FCER1G', 'IGJ', 'CST3',
# 'LGALS2', 'GZMA', 'PRF1', 'HIST1H2AC', 'FCER1A', 'STK17A', 'HLA-DQB1',
# 'CST7', 'SNRPB', 'LINC00926', 'HLA-DPA1', 'MS4A6A', 'UBE2L6', 'ATP6V1F',
# 'SF3B5', 'FYB', 'C9orf142', 'APOBEC3A', 'IGLL5', 'NPC2', 'GRN', 'LYAR',
# 'PYCARD', 'LGALS1', 'PF4', 'PTCRA', 'IFITM3']
# Probeset (selected with Spapros, see basic selection tutorial)
probeset = [
'PF4', 'HLA-DPB1', 'FCGR3A', 'GZMB', 'CCL5', 'S100A8', 'IL32', 'HLA-DQA1', 'NKG7', 'AIF1', 'CD79A', 'LTB', 'TYROBP',
'HLA-DMA', 'GZMK', 'HLA-DRB1', 'FCN1', 'S100A11', 'GNLY', 'GZMH'
]
# Reference probesets
reference_sets = sp.se.select_reference_probesets(adata, n=20)
Reference probeset selection.............................. ━━━━━━━━━━━━━━━━━━━━ 0/4 0:00:00
Evaluate sets
[12]:
evaluator = sp.ev.ProbesetEvaluator(adata, verbosity=2, results_dir=None)
[13]:
evaluator.evaluate_probeset(probeset, set_id="Spapros")
SPAPROS PROBESET EVALUATION: Shared metric computations................................ ━━━━━━━━━━━━━━━━━━━━ 3/3 0:00:02 Computing shared compuations for knn_overlap............ ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:01 Computing shared compuations for gene_corr.............. ━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 Probeset specific pre computations........................ ━━━━━━━━━━━━━━━━━━━━ 3/3 0:00:02 Computing pre compuations for knn_overlap............... ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:02 Final probeset specific computations...................... ━━━━━━╸━━━━━━━━━━━━━ 1/3 0:00:04 Computing final compuations for knn_overlap............. ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:00 Computing final compuations for forest_clfs............. ━━━━━━━━━━━━╸━━━━━━━ 16/25 0:00:03
[14]:
for set_id, df in reference_sets.items():
gene_set = df[df["selection"]].index.to_list()
evaluator.evaluate_probeset(gene_set, set_id=set_id)
SPAPROS PROBESET EVALUATION: Shared metric computations................................ ━━━━━━━━━━━━━━━━━━━━ 3/3 0:00:02 Computing shared compuations for knn_overlap............ ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:01 Computing shared compuations for gene_corr.............. ━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 Probeset specific pre computations........................ ━━━━━━━━━━━━━━━━━━━━ 3/3 0:00:01 Computing pre compuations for knn_overlap............... ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:01 Final probeset specific computations...................... ━━━━━━━━━━━━━━━━━━━━ 3/3 0:00:05 Computing final compuations for knn_overlap............. ━━━━━━━━━━━━━━━━━━━━ 6/6 0:00:00 Computing final compuations for forest_clfs............. ━━━━━━━━━━━━━━━━━━━━ 25/25 0:00:05 Computing final compuations for gene_corr............... ━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 FINISHED
[15]:
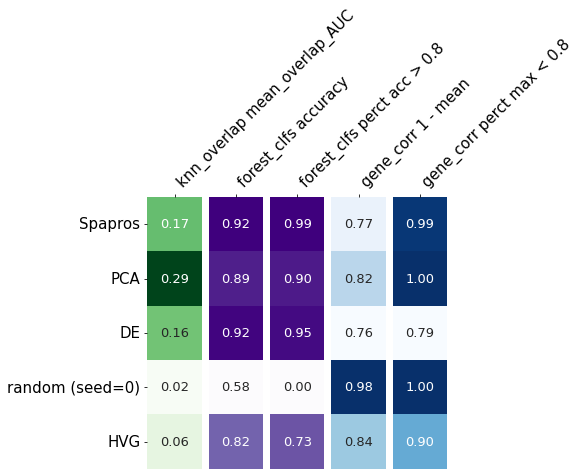
evaluator.summary_results#.index.tolist()
[15]:
| knn_overlap mean_overlap_AUC | forest_clfs accuracy | forest_clfs perct acc > 0.8 | gene_corr 1 - mean | gene_corr perct max < 0.8 | |
|---|---|---|---|---|---|
| Spapros | 0.167580 | 0.924543 | 0.985218 | 0.772479 | 0.993647 |
| PCA | 0.293958 | 0.889088 | 0.902960 | 0.820372 | 1.000000 |
| DE | 0.160032 | 0.919459 | 0.953571 | 0.757868 | 0.792178 |
| random (seed=0) | 0.023879 | 0.575304 | 0.000000 | 0.976067 | 1.000000 |
| HVG | 0.056358 | 0.818277 | 0.729101 | 0.840755 | 0.900000 |
Visualize the results
[9]:
import matplotlib as mpl
mpl.rcParams['figure.dpi'] = 60
Note: After running the spapros evaluation pipeline, the results are stored in a directory, that is by default called probeset_evaluation. If you run another evaluation after this tutorial, be careful that you either specify another dir or delete the previously created results directory because otherwise parts will be overwritten and parts will be falsely reused! If you do not want to save results, initialize the ProbesetEvaluator with dir=None, like in this tutorial.