spapros.se.ProbesetSelector.plot_clf_genes
- ProbesetSelector.plot_clf_genes(basis='X_umap', celltypes=None, till_rank=1, importance_th=None, add_marker_genes=True, neighbors_params={}, umap_params={}, **kwargs)
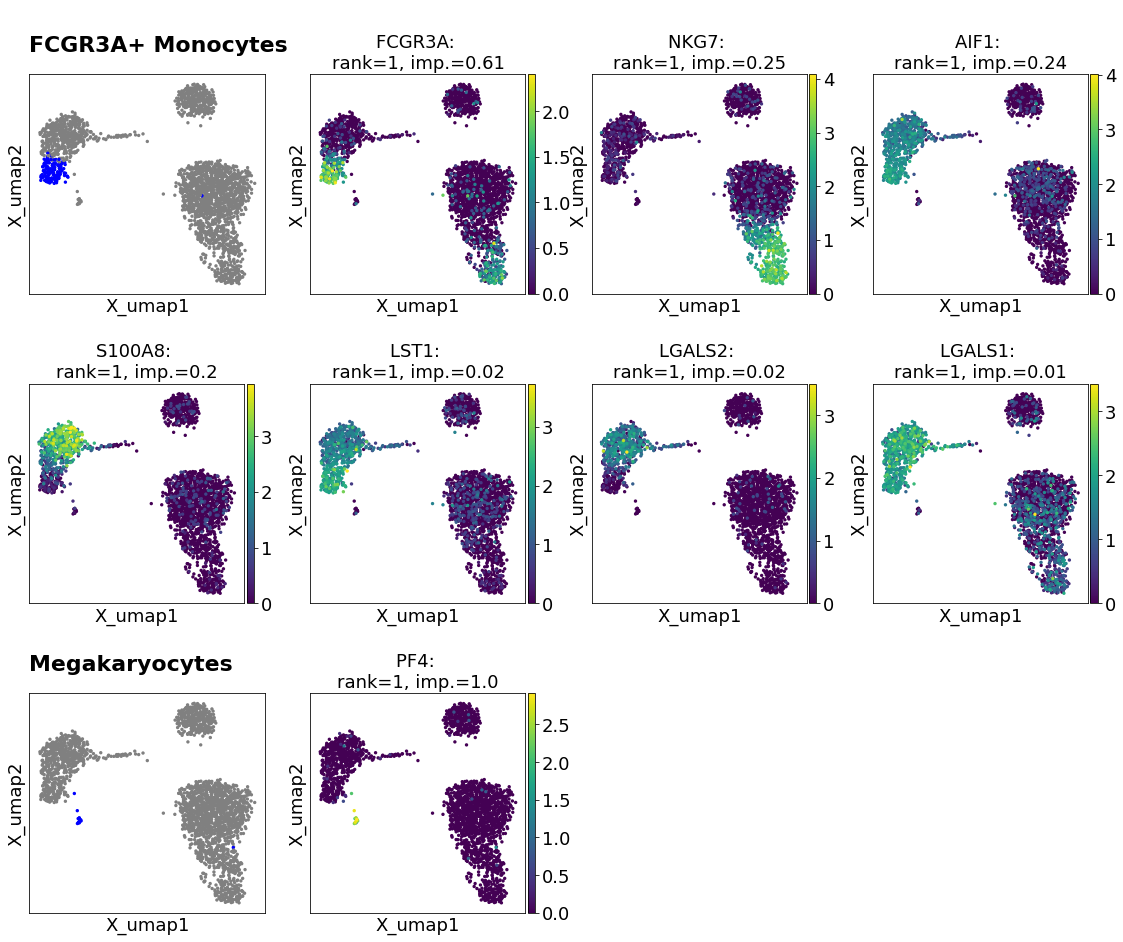
Plot umaps of selected genes needed for cell type classification of each cell type.
- Parameters:
basis (str) – Name of the
obsmembedding to use.celltypes (Optional[List[str]]) – Subset of cell types for which to plot decision genes. If None,
celltypesis used.till_rank (Optional[int]) – Plot decision genes only up to the given tree rank of the probeset list.
importance_th (Optional[float]) – Only plot genes with a tree feature importance above the given threshold.
add_marker_genes (bool) – Whether to add subplots for marker genes from
marker_listfor each celltype. TODO: what about cell types that only occur in the marker list?neighbors_params (dict) – Parameters for
sc.pp.neighbors(). Only applicable ifadata.obsm[basis]does not exist. TODO: do we rly need that parameter? Would be fine to always expect a pre calculated embedding!umap_params (dict) – Parameters for
sc.tl.umap(). Only applicable ifadata.obsm[basis]does not exist. TODO: do we rly need that parameter? Would be fine to always expect a pre calculated embedding!kwargs – Keyword arguments of
selection_histogram().
Example
(Takes a few minutes to calculate)
import spapros as sp adata = sp.ut.get_processed_pbmc_data() selector = sp.se.ProbesetSelector(adata, "celltype", n=30, verbosity=0) selector.select_probeset() selector.plot_clf_genes(n_cols=4,celltypes=["FCGR3A+ Monocytes","Megakaryocytes"])
TODO: this function and pl.clf_genes_umaps need to be tested on all argument combinations + can be optimized.