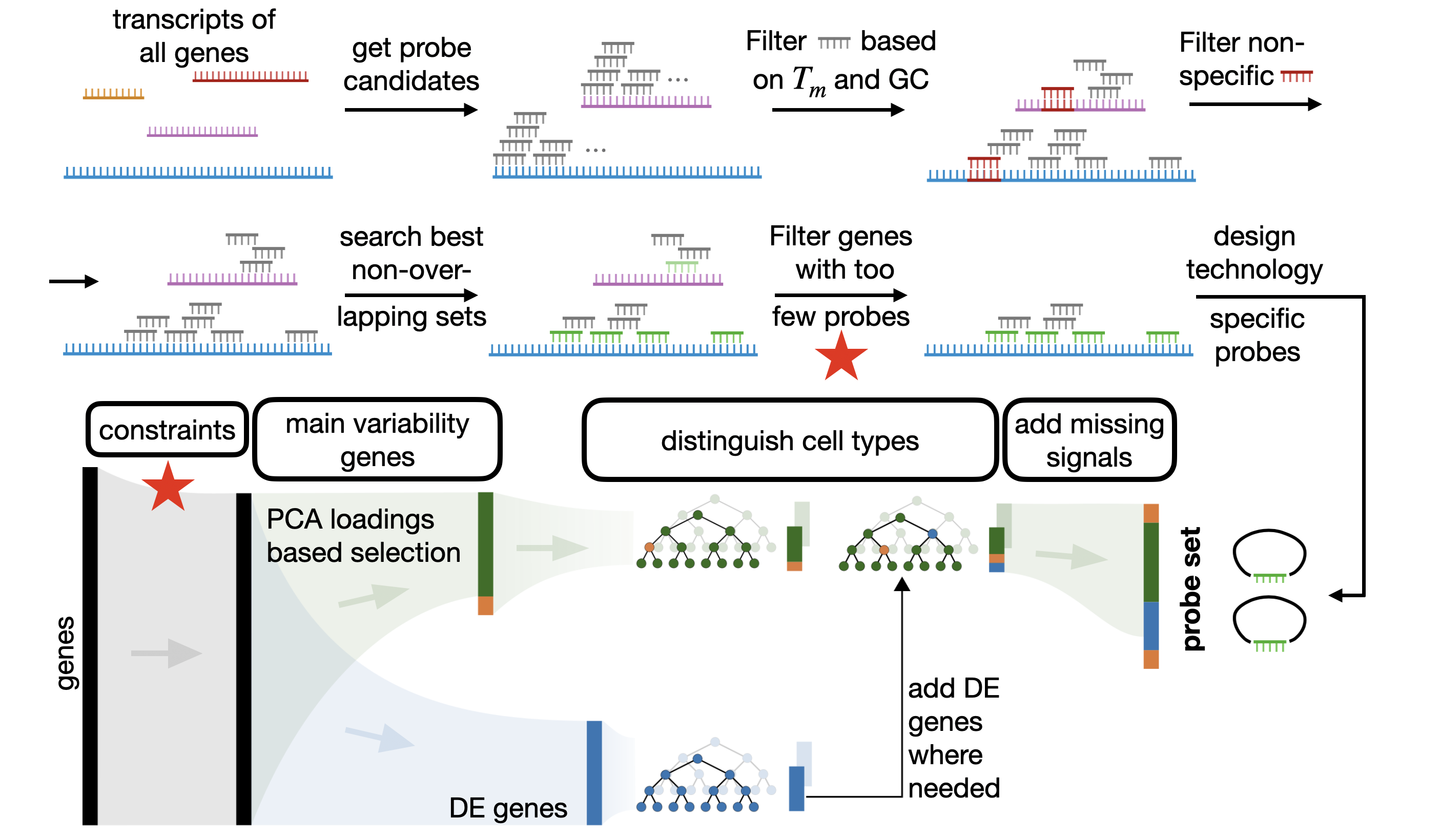
End to end selection
In this tutorial we showcase how to design probesets and select a suitable gene set with the spapros package. For all genes we design probes that fulfill experiment specific requirements and select only genes for which we can design sufficient probes. Spapros then selects genes that can distinguish the cell types in the data set and capture transcriptomic varation beyond cell type labels. The final probe sequences are designed in a last step for all selected genes. The figure below gives and
overview on the pipeline.
[2]:
Import Packages
Besides spapros also install oligo_designer_toolsuite if not done already. First we need to install some dependencies:
conda config --add channels bioconda
conda config --add channels conda-forge
conda update conda
conda update --all
conda install "blast>=2.12"
conda install "bedtools>=2.30"
conda install "bowtie>=1.3.1"
conda install "bowtie2>=2.5"
To run the code below we need to install the current dev version of the oligo designer:
git clone https://github.com/HelmholtzAI-Consultants-Munich/oligo-designer-toolsuite.git
cd oligo-designer-toolsuite
git switch pipelines
pip install -e .
Otherwise, if that didn’t work, try:
pip install oligo_designer_toolsuite
[3]:
import scanpy as sc
sc.settings.verbosity = 0
sc.logging.print_header()
import spapros as sp
print(f"spapros=={sp.__version__}")
from oligo_designer_toolsuite.pipelines import ScrinshotProbeDesigner #, MerfishProbeDesigner, SeqfishPlusProbeDesigner
scanpy==1.9.3 anndata==0.9.2 umap==0.5.3 numpy==1.24.4 scipy==1.11.1 pandas==1.5.3 scikit-learn==1.3.0 statsmodels==0.14.0 python-igraph==0.9.11 pynndescent==0.5.10
spapros==0.1.3
Load and Preprocess Data
For this tutorial, we use a PBMC example scRNA-seq reference dataset. The count data should be log-normalised and genes should not be scaled to mean=0 and std=1. We can load the processed version of the data, including cell / gene filters, cell type annotations, and the umap embedding, directly with sp.ut.get_processed_pbmc_data() function. For a step by step processing of the PBMC dataset please refer to the basic usage tutorial. For sake
of simplicity, we pre-select the top 1000 highly variable genes for the probe and geneset selection. In real world applications we typically go for top 8000 genes.
[5]:
pbmc_data = sp.ut.get_processed_pbmc_data(n_hvg=1000)
highly_variable_genes = sorted(pbmc_data.var.loc[pbmc_data.var['highly_variable']].index.tolist())
print(f"Number of highly variable genes: {len(highly_variable_genes)}")
Number of highly variable genes: 1000
Probeset Design
Before choosing a gene panel, we design probesets for our given set of 1000 highly variable genes that fulfill certain experiment-specific criteria. Therefore, we first create an instance of a ProbeDesigner class, where we can choose from ScrinshotProbeDesigner, MerfishProbeDesigner an SeqfishPlusProbeDesigner (see our resource
table for an overview of differences between the technologies). For each of those classes, we need to define an output directory and set the parameters write_removed_genes (if true, save gene with insufficient probes in a file) and write_intermediate_steps (if true, save the probe
database after each processing step, such that the pipline can resume from a certain step onwards).
Here, we showcase the probe design of padlock probes.
[6]:
probe_designer = ScrinshotProbeDesigner(dir_output="./output")
2023-08-22 17:25:41,843 [INFO] Parameters Init:
2023-08-22 17:25:41,844 [INFO] dir_output = ./output
2023-08-22 17:25:41,845 [INFO] write_removed_genes = True
2023-08-22 17:25:41,846 [INFO] write_intermediate_steps = True
After instatiating a ProbeDesigner class, we need to load the annotation we are using. Our example dataset uses the NCBI gene annotation. Hence, we define ncbi as source and define the NCBI-specific parameters taxon, species and annotation_release. Apart from NCBI annotation, we can also choose an Ensembl annotation. If source="ncbi" or source="ensembl" is choosen, the annotation files are automatically downloaded from their servers. In addition, we can provide a custom
annotation when specifying source="custom".
Parameters for annotation loader
source: define annotation source -> currently supported: ncbi, ensembl and custom
NCBI annnotation parameters: - taxon: taxon of the species, valid taxa are: archaea, bacteria, fungi, invertebrate, mitochondrion, plant, plasmid, plastid, protozoa, vertebrate_mammalian, vertebrate_other, viral - species: species name in NCBI download format, e.g. ‘Homo_sapiens’ for human; see here for available species name - annotation_release: release number (e.g. 109 or 109.20211119 for ncbi) of annotation or ‘current’ to use
most recent annotation release. Check out release numbers for NCBI at ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/annotation/annotation_releases/
Ensembl annotation parameters: - species: species name in ensembl download format, e.g. ‘homo_sapiens’ for human; see http://ftp.ensembl.org/pub/release-108/gtf/ for available species names - annotation_release: release number of annotation, e.g. ‘release-108’ or ‘current’ to use most recent annotation release. Check out release numbers for ensemble at ftp.ensembl.org/pub/
Custom annotation parameters: - file_annotation: GTF file with gene annotation - file_sequence: FASTA file with genome sequence - files_source: original source of the genomic files -> optional - species: species of provided annotation, leave empty if unknown -> optional - annotation_release: release number of provided annotation, leave empty if unknown -> optional - genome_assembly: genome assembly of provided annotation, leave empty if unknown -> optional
[6]:
# example for ncbi annotation loader
source = "ncbi"
params = {
"taxon": "vertebrate_mammalian",
"species": "Homo_sapiens",
"annotation_release": "110",
}
# example for ensembl annotation loader
# source = "ensembl"
# params = {
# "species": "homo_sapiens",
# "annotation_release": "109",
# }
# example for custom annotation loader
# source = "custom"
# params = {
# "file_annotation": "./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.gtf",
# "file_sequence": "./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.fna",
# "files_source": "NCBI",
# "species": "Homo_sapiens",
# "annotation_release": "110",
# "genome_assembly": "GRCh38.p14",
# }
probe_designer.load_annotations(source=source, source_params=params)
2023-05-26 17:13:37,570 [INFO] Parameters Load Annotations:
2023-05-26 17:13:37,572 [INFO] source = custom
2023-05-26 17:13:37,573 [INFO] source_params = {'file_annotation': './output/annotation/GCF_000001405.40_GRCh38.p14_genomic.gtf', 'file_sequence': './output/annotation/GCF_000001405.40_GRCh38.p14_genomic.fna', 'files_source': 'NCBI', 'species': 'Homo_sapiens', 'annotation_release': '110', 'genome_assembly': 'GRCh38.p14'}
2023-05-26 17:16:31,649 [INFO] The following annotation files are used for GTF annotation of regions: ./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.gtf and for fasta sequence file: ./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.fna .
2023-05-26 17:16:31,653 [INFO] The annotations are from NCBI source, for the species: Homo_sapiens, release number: 110 and genome assembly: GRCh38.p14
After downloading the annotations, we have to create the oligo database. Running the function below, will automatically create a transcriptome from the given annotation (therefore, the provided GTF file must contain transcript and exon information) and use this transcriptome to create all possible probes for each gene, that is provided in the gene list.
Parameters for Probe Sequences Database
probe_length_min: minimum length of probesprobe_length_max: maximum length of probesmin_probes_per_gene: minimum number of probes that a gene must have before it gets deletedregion: Target sequence type for which probes are designed (choose from: “transcript”, “genome”, “cds”)
load_probe_database(). See example code in the cells below (commented).[7]:
probe_length_min = 38
probe_length_max = 45
min_probes_per_gene = 3
region = "transcript"
# highly_variable_genes = highly_variable_genes[100:]
probe_database, file_database = probe_designer.create_probe_database(genes=highly_variable_genes, probe_length_min=probe_length_min, probe_length_max=probe_length_max, region=region, min_probes_per_gene=min_probes_per_gene, n_jobs=4)
2023-05-26 17:16:31,766 [INFO] Parameters Create Database:
2023-05-26 17:16:31,767 [INFO] genes = ['AAGAB', 'AATF', 'ABCC10', 'ABHD12', 'ABHD17B', 'ABHD5', 'ABRACL', 'ABT1', 'AC005082.12', 'AC074138.3', 'AC093323.3', 'ACAP1', 'ACBD3', 'ACD', 'ACOT13', 'ACP1', 'ACRBP', 'ACTL6A', 'ACTR6', 'ACVR2A', 'ADAL', 'ADAM10', 'ADAM28', 'ADD1', 'ADIPOR2', 'ADPRM', 'ADSL', 'AEBP1', 'AGPAT1', 'AHSA1', 'AIF1', 'AIM2', 'AKTIP', 'AL928768.3', 'ALKBH7', 'ANAPC13', 'ANKAR', 'ANKEF1', 'ANKRD27', 'ANKRD54', 'AP001462.6', 'AP003419.16', 'AP3M2', 'AP4B1-AS1', 'AP4S1', 'APOBEC3A', 'APOBEC3B', 'APOBEC3G', 'AQP3', 'ARHGAP11A', 'ARHGAP19', 'ARHGAP24', 'ARHGAP33', 'ARHGAP6', 'ARID4A', 'ARIH2OS', 'ARL2', 'ARL2BP', 'ARL4A', 'ARL6IP5', 'ARMC7', 'ARMCX5', 'ARRDC3', 'ARRDC4', 'ARSD', 'ARSG', 'ARVCF', 'ASB8', 'ASXL2', 'ATAD3C', 'ATF7IP2', 'ATG16L1', 'ATP10A', 'ATP5H', 'ATP5O', 'ATP5SL', 'ATP6V0E2', 'ATXN1L', 'ATXN3', 'AURKC', 'BABAM1', 'BACE2', 'BAZ2A', 'BBX', 'BCDIN3D', 'BET1', 'BEX4', 'BGLAP', 'BLNK', 'BLZF1', 'BMPR2', 'BNIP2', 'BOLA1', 'BOLA3', 'BRAT1', 'BRWD1', 'BTN3A1', 'BTN3A2', 'BUB3', 'C10orf32', 'C12orf45', 'C14orf1', 'C14orf166', 'C14orf80', 'C15orf57', 'C16orf13', 'C16orf52', 'C16orf54', 'C16orf58', 'C16orf74', 'C16orf80', 'C17orf59', 'C17orf62', 'C19orf33', 'C19orf52', 'C1QA', 'C1QB', 'C1QC', 'C1orf162', 'C1orf35', 'C21orf33', 'C2CD4D', 'C2orf76', 'C2orf88', 'C3orf18', 'C5orf15', 'C5orf42', 'C8orf44', 'C9orf142', 'C9orf16', 'C9orf37', 'CAMK1D', 'CAMK2G', 'CAMK2N1', 'CAPN12', 'CARHSP1', 'CARS', 'CASC4', 'CBX5', 'CCDC115', 'CCDC122', 'CCDC66', 'CCDC91', 'CCL3', 'CCL4', 'CCL5', 'CCND2', 'CCNG1', 'CCP110', 'CCT4', 'CCT7', 'CD160', 'CD19', 'CD2', 'CD247', 'CD274', 'CD2AP', 'CD320', 'CD72', 'CD79A', 'CD79B', 'CD82', 'CD9', 'CD96', 'CDC123', 'CDC16', 'CDC37', 'CDC40', 'CDK19', 'CDKN2A', 'CEACAM4', 'CEBPB', 'CECR5', 'CEP120', 'CEP68', 'CEP85L', 'CEPT1', 'CES4A', 'CGRRF1', 'CHD2', 'CHD7', 'CHERP', 'CHI3L2', 'CHPF2', 'CIAPIN1', 'CISD1', 'CISH', 'CITED4', 'CKS1B', 'CLDN5', 'CLEC2B', 'CLIC3', 'CLNS1A', 'CLPX', 'CLU', 'CLYBL', 'CMTM5', 'CNEP1R1', 'COMMD10', 'COQ7', 'CORO1B', 'COTL1', 'CPNE2', 'CPQ', 'CPSF3L', 'CR1', 'CRIP3', 'CRTC2', 'CST3', 'CST7', 'CTA-29F11.1', 'CTB-113I20.2', 'CTB-152G17.6', 'CTC-444N24.11', 'CTD-2015H6.3', 'CTD-2302E22.4', 'CTD-2368P22.1', 'CTD-2537I9.12', 'CTSS', 'CTSW', 'CWC15', 'CWC27', 'CXCL10', 'CXCL3', 'CYB5B', 'CYTH2', 'DAGLB', 'DCAF5', 'DDI2', 'DDT', 'DDX1', 'DDX17', 'DDX46', 'DDX56', 'DENND1C', 'DENND2D', 'DENND5B', 'DENND6A', 'DERL1', 'DEXI', 'DHX34', 'DHX9', 'DIDO1', 'DIMT1', 'DIS3', 'DISP1', 'DLST', 'DMTN', 'DNAJA3', 'DNAJB14', 'DNAJC10', 'DNAJC15', 'DNAJC2', 'DNAJC27', 'DNASE1L3', 'DNMT3A', 'DOK3', 'DPH6', 'DPY19L4', 'DRAXIN', 'DSCR3', 'DTX3', 'DUS3L', 'DUSP10', 'EAF2', 'EARS2', 'ECHDC1', 'EDC3', 'EID2', 'EIF1AY', 'EIF1B', 'EIF2B1', 'EIF3D', 'ELANE', 'ELOF1', 'ELOVL4', 'ELP6', 'EMB', 'EMG1', 'EML6', 'ENTPD3-AS1', 'EOGT', 'ERH', 'ERV3-1', 'EVA1B', 'EWSR1', 'EXOC6', 'F5', 'FADS1', 'FAM107B', 'FAM173A', 'FAM210B', 'FAM96A', 'FAM98A', 'FBXL14', 'FBXO21', 'FBXO33', 'FBXO4', 'FBXW4', 'FCER1A', 'FCER1G', 'FCGR2B', 'FCGR3A', 'FCN1', 'FCRLA', 'FEM1A', 'FERMT3', 'FGFBP2', 'FH', 'FHL1', 'FKBP3', 'FKBP5', 'FLOT1', 'FMO4', 'FN3KRP', 'FNBP4', 'FNTA', 'FOPNL', 'FRY-AS1', 'FUS', 'FXN', 'FYB', 'G0S2', 'GADD45B', 'GALT', 'GBGT1', 'GBP1', 'GDF11', 'GFER', 'GGA3', 'GGNBP2', 'GIMAP2', 'GIMAP4', 'GIMAP5', 'GIMAP7', 'GIT2', 'GMPPA', 'GNE', 'GNG11', 'GNG3', 'GNLY', 'GNPAT', 'GOLGB1', 'GP9', 'GPATCH4', 'GPKOW', 'GPR171', 'GPR183', 'GPR35', 'GPS1', 'GPX1', 'GRAP', 'GRN', 'GSTP1', 'GTPBP6', 'GUSB', 'GYS1', 'GZMA', 'GZMB', 'GZMH', 'GZMK', 'HAGH', 'HBA1', 'HBP1', 'HCFC2', 'HDAC1', 'HDAC5', 'HDAC9', 'HELQ', 'HEMK1', 'HERPUD2', 'HIST1H1B', 'HIST1H2AC', 'HIST1H2AH', 'HLA-DMA', 'HLA-DMB', 'HLA-DOB', 'HLA-DPA1', 'HLA-DPB1', 'HLA-DQA1', 'HLA-DQB1', 'HLA-DRB1', 'HMBOX1', 'HMGCL', 'HMGXB4', 'HNRNPH3', 'HOOK2', 'HOPX', 'HSPB11', 'HVCN1', 'ICAM2', 'ICOS', 'ICOSLG', 'ID2', 'IDUA', 'IFFO1', 'IFI27', 'IFIT1', 'IFIT2', 'IFITM3', 'IGFBP7', 'IGJ', 'IGLL5', 'IL1B', 'IL1RAP', 'IL23A', 'IL24', 'IL27RA', 'IL32', 'IL6', 'IL8', 'ILF3', 'ILF3-AS1', 'ING5', 'INSL3', 'INTS12', 'INTS2', 'IP6K1', 'IQCE', 'IRF8', 'IRF9', 'ISCA2', 'ISOC1', 'ITGA2B', 'ITGB7', 'ITM2A', 'ITSN2', 'JAKMIP1', 'JUND', 'KARS', 'KCNG1', 'KCNQ1OT1', 'KIAA0040', 'KIAA0125', 'KIAA0196', 'KIAA1430', 'KIF3A', 'KIF3C', 'KIF5B', 'KLHL24', 'KLRB1', 'KLRG1', 'KRBOX4', 'LAMP3', 'LARS', 'LAT2', 'LBR', 'LDLRAP1', 'LGALS1', 'LGALS2', 'LGALS3', 'LILRA4', 'LIN52', 'LINC00494', 'LINC00662', 'LINC00886', 'LINC00926', 'LINC00936', 'LINC01013', 'LIX1L', 'LONRF1', 'LPIN1', 'LRBA', 'LRRIQ3', 'LSM14A', 'LST1', 'LTB', 'LTV1', 'LUC7L', 'LUC7L3', 'LYAR', 'LYPD2', 'LYPLA1', 'LYRM4', 'LYSMD4', 'LZTS2', 'MADD', 'MAEA', 'MAGEH1', 'MAL', 'MALT1', 'MAP2K7', 'MARCKSL1', 'MCF2L', 'MCM3', 'MDS2', 'MED30', 'MED9', 'METTL21A', 'METTL3', 'METTL8', 'MFF', 'MFSD10', 'MIS18A', 'MKKS', 'MLLT11', 'MLLT6', 'MMADHC', 'MMP9', 'MNAT1', 'MOCS2', 'MORF4L2', 'MPHOSPH10', 'MRM1', 'MRPL1', 'MRPL19', 'MRPL42', 'MRPS12', 'MRPS33', 'MS4A1', 'MS4A6A', 'MTERFD2', 'MTIF2', 'MTRF1', 'MUM1', 'MYADM', 'MYCBP2', 'MYL9', 'MYO1E', 'MYOM2', 'MZB1', 'MZT1', 'NAA20', 'NAP1L4', 'NAPA-AS1', 'NARG2', 'NAT9', 'NBR1', 'NCOR2', 'NCR3', 'NDUFA10', 'NDUFA12', 'NECAB3', 'NEFH', 'NEK8', 'NELFB', 'NEMF', 'NFAT5', 'NFE2L2', 'NFIC', 'NFU1', 'NIT2', 'NKAP', 'NKG7', 'NKTR', 'NME3', 'NME6', 'NMNAT3', 'NNT-AS1', 'NOC4L', 'NOG', 'NOL11', 'NONO', 'NOP58', 'NPC2', 'NPHP3', 'NPRL2', 'NR2C1', 'NR3C1', 'NSA2', 'NT5C', 'NT5C3A', 'NUDCD1', 'NUDT16L1', 'NUP54', 'NXT2', 'OARD1', 'OAT', 'OBSCN', 'ODC1', 'ORAI1', 'ORC2', 'OSBPL1A', 'OSBPL7', 'OXLD1', 'P2RX5', 'P2RY10', 'PACS1', 'PACSIN2', 'PAICS', 'PARP1', 'PARS2', 'PASK', 'PAWR', 'PAXIP1-AS1', 'PBLD', 'PBRM1', 'PCNA', 'PCSK7', 'PDCD1', 'PDCD2L', 'PDE6B', 'PDIA3', 'PDIK1L', 'PDK2', 'PDXDC1', 'PDZD4', 'PEMT', 'PEX16', 'PEX26', 'PF4', 'PGM1', 'PGM2L1', 'PHACTR4', 'PHF12', 'PHF14', 'PHF3', 'PIGF', 'PIGU', 'PIGX', 'PIK3R1', 'PITHD1', 'PITPNA-AS1', 'PJA1', 'PKIG', 'PLA2G12A', 'PLCL1', 'PLD6', 'PLEKHA1', 'PLEKHA3', 'PLRG1', 'PMEPA1', 'PNOC', 'POLR2I', 'POLR2K', 'POLR3E', 'POMT1', 'PPA2', 'PPBP', 'PPIE', 'PPIG', 'PPIL2', 'PPIL4', 'PPP1R14A', 'PPP1R2', 'PPP2R1B', 'PPP6C', 'PPT2-EGFL8', 'PQBP1', 'PRAF2', 'PRDX1', 'PRELID2', 'PRF1', 'PRICKLE1', 'PRKACB', 'PRKCB', 'PRKD2', 'PRMT2', 'PRNP', 'PRPF31', 'PRPS2', 'PRR5', 'PSMD14', 'PTCRA', 'PTGDR', 'PTGDS', 'PTGES2', 'PTPN7', 'PURA', 'PWP1', 'PXMP4', 'PYCARD', 'R3HDM1', 'R3HDM2', 'RAB40C', 'RABEP2', 'RABL6', 'RAD51B', 'RALBP1', 'RALY', 'RASD1', 'RASGRP2', 'RBM25', 'RBM26-AS1', 'RBM39', 'RBM4', 'RBM48', 'RBM5', 'RBM7', 'RBPJ', 'RCE1', 'RCHY1', 'RCL1', 'RCN2', 'RDH14', 'RELB', 'REXO2', 'RFC1', 'RFC5', 'RFNG', 'RFPL2', 'RGS14', 'RIC3', 'RIOK1', 'RIOK2', 'RNF113A', 'RNF125', 'RNF139', 'RNF14', 'RNF168', 'RNF187', 'RNF213', 'RNF25', 'RNF26', 'RORA', 'RP1-28O10.1', 'RP11-1055B8.7', 'RP11-138A9.2', 'RP11-141B14.1', 'RP11-142C4.6', 'RP11-162G10.5', 'RP11-164H13.1', 'RP11-178G16.4', 'RP11-18H21.1', 'RP11-211G3.2', 'RP11-219B17.1', 'RP11-219B4.7', 'RP11-252A24.3', 'RP11-291B21.2', 'RP11-314N13.3', 'RP11-324I22.4', 'RP11-349A22.5', 'RP11-378J18.3', 'RP11-390B4.5', 'RP11-398C13.6', 'RP11-400F19.6', 'RP11-421L21.3', 'RP11-428G5.5', 'RP11-432I5.1', 'RP11-468E2.4', 'RP11-488C13.5', 'RP11-493L12.4', 'RP11-527L4.5', 'RP11-545I5.3', 'RP11-589C21.6', 'RP11-5C23.1', 'RP11-701P16.5', 'RP11-706O15.1', 'RP11-70P17.1', 'RP11-727F15.9', 'RP11-798G7.6', 'RP11-879F14.2', 'RP11-950C14.3', 'RP3-325F22.5', 'RP5-1073O3.7', 'RP5-827C21.4', 'RP5-887A10.1', 'RPH3A', 'RPL39L', 'RPL7L1', 'RPN2', 'RPS6KL1', 'RPUSD2', 'RRAGC', 'RRS1', 'RUNDC1', 'S100A11', 'S100A12', 'S100A8', 'S100B', 'SAFB2', 'SAMD1', 'SAMD3', 'SAMSN1', 'SARDH', 'SARS', 'SAT1', 'SCAI', 'SCAPER', 'SCGB3A1', 'SCPEP1', 'SDCCAG8', 'SDPR', 'SEC61A2', 'SELL', 'SEPT11', 'SERAC1', 'SETD1B', 'SF3B1', 'SF3B5', 'SH3GLB1', 'SH3KBP1', 'SHOC2', 'SHPK-1', 'SIAH2', 'SIRPG', 'SIRT1', 'SIVA1', 'SLA', 'SLBP', 'SLC22A4', 'SLC25A11', 'SLC25A12', 'SLC25A14', 'SLC27A1', 'SLC2A13', 'SLC35A2', 'SLC48A1', 'SLFN5', 'SMARCA4', 'SMARCC2', 'SMC2', 'SMCHD1', 'SMDT1', 'SMIM14', 'SMIM7', 'SNAP47', 'SNHG12', 'SNHG8', 'SNTA1', 'SNX29P2', 'SOX13', 'SPARC', 'SPATA7', 'SPG7', 'SPIB', 'SPIN1', 'SPOCD1', 'SPON2', 'SPSB2', 'SREBF1', 'SRM', 'SRP9', 'SRSF6', 'SSBP1', 'ST3GAL2', 'STAMBP', 'STAU2', 'STK17A', 'STK38', 'STMN1', 'STOML2', 'STUB1', 'STX16', 'STX18', 'SUCLG2', 'SUOX', 'SURF1', 'SURF6', 'SWAP70', 'SYCE1', 'SYP', 'SYVN1', 'TACR2', 'TADA2A', 'TAF10', 'TAF12', 'TAF1D', 'TAL1', 'TALDO1', 'TAPBP', 'TARSL2', 'TASP1', 'TBC1D15', 'TBCK', 'TBXA2R', 'TCEAL4', 'TCEAL8', 'TCL1A', 'TCL1B', 'TCP1', 'TDG', 'TERF2IP', 'TGFBRAP1', 'THAP2', 'THEM4', 'THOC7', 'THUMPD3', 'THYN1', 'TIGIT', 'TIMM10B', 'TMEM116', 'TMEM138', 'TMEM140', 'TMEM14B', 'TMEM165', 'TMEM177', 'TMEM194A', 'TMEM219', 'TMEM242', 'TMEM40', 'TMEM60', 'TMEM80', 'TMEM87A', 'TMEM87B', 'TMEM91', 'TMTC2', 'TMX2', 'TMX3', 'TNFRSF17', 'TNFRSF25', 'TNFRSF4', 'TNFRSF9', 'TNFSF10', 'TOP1MT', 'TOP2B', 'TRABD2A', 'TRAF3IP3', 'TRAPPC12-AS1', 'TRAPPC3', 'TREML1', 'TRIM23', 'TRIP12', 'TRIT1', 'TRMT61A', 'TRPM4', 'TSC22D1', 'TSPAN15', 'TSSC1', 'TTC1', 'TTC14', 'TTC3', 'TTC8', 'TTN-AS1', 'TUBB1', 'TUBG2', 'TYMP', 'TYROBP', 'U2SURP', 'UBA5', 'UBAC2', 'UBE2D2', 'UBE2D4', 'UBE2K', 'UBE2Q1', 'UBE3A', 'UBIAD1', 'UBLCP1', 'UBXN4', 'UCK1', 'UNC45A', 'UQCC1', 'URB2', 'URGCP', 'USP30', 'USP33', 'USP36', 'USP38', 'USP5', 'USP7', 'VAMP5', 'VDAC3', 'VIPR1', 'VPS13A', 'VPS13C', 'VPS25', 'VPS26B', 'VPS28', 'VTI1A', 'VTI1B', 'WARS2', 'WBP2NL', 'WDR55', 'WDR91', 'WDYHV1', 'WNK1', 'WTAP', 'XCL2', 'XPOT', 'XRRA1', 'XXbac-BPG299F13.17', 'YEATS2', 'YES1', 'YPEL2', 'YPEL3', 'YTHDF2', 'ZAP70', 'ZBED5-AS1', 'ZBP1', 'ZC3H15', 'ZCCHC11', 'ZCCHC9', 'ZFAND4', 'ZNF175', 'ZNF232', 'ZNF256', 'ZNF263', 'ZNF276', 'ZNF32', 'ZNF350', 'ZNF436', 'ZNF45', 'ZNF493', 'ZNF503', 'ZNF528', 'ZNF559', 'ZNF561', 'ZNF587B', 'ZNF594', 'ZNF653', 'ZNF682', 'ZNF688', 'ZNF718', 'ZNF747', 'ZNF799', 'ZNF836', 'ZNF92', 'ZRANB3', 'ZSWIM6', 'ZUFSP']
2023-05-26 17:16:31,768 [INFO] probe_length_min = 38
2023-05-26 17:16:31,769 [INFO] probe_length_max = 45
2023-05-26 17:16:31,771 [INFO] min_probes_per_gene = 3
2023-05-26 17:16:31,772 [INFO] n_jobs = 4
2023-05-26 17:56:26,665 [INFO] Genes with <= 3 probes will be removed from the probe database and their names will be stored in './output/regions_with_insufficient_oligos.txt'.
2023-05-26 17:56:26,862 [INFO] Step - Generate Probes: the database contains 35957196 probes from 887 genes.
In order to create experiment-specific probes, we have to apply several filter to each probe, e.g. melting temperature or GC content filters.
Parameters for Property Filters
Parameters for Probe Sequence: - GC_content_min: minimum GC content of probes - GC_content_max: maximum GC content of probes - Tm_min: minimum melting temperature of probes - Tm_max: maximum melting temperature of probes
Parameters for Padlock Arms: - min_arm_length: minimum length of each arm - max_arm_Tm_dif: maximum melting temperature difference of both arms - arm_Tm_min: minimum melting temperature of each arm (difference shouldn’t be higher than 5! But range is not super important, the lower the better) - arm_Tm_max: maximum melting temperature of each arm
Parameters for Melting Temperature: - Tm_parameters_probe: melting temperature parameters for probe design - Tm_chem_correction_param_pobe: parameters for chemical correction of melting temperature for probe design
Note: The melting temperature is used in 2 different stages (probe and detection oligo design), where a few parameters are shared and the others differ. Parameters for melting temperature -> for more information on parameters, see:here
[8]:
####### Load existing database #######
# file_database = "./output/oligo_database/probe_database_initial.txt"
# min_probes_per_gene = 3
# probe_database = probe_designer.load_probe_database(file_database=file_database, min_probes_per_gene=min_probes_per_gene)
####### Apply Property Filter #######
GC_content_min=40
GC_content_max=60
Tm_min=52
Tm_max=67
min_arm_length=10
max_arm_Tm_dif=2
arm_Tm_min=38
arm_Tm_max=49
probe_database, file_database = probe_designer.filter_probes_by_property(probe_database, GC_content_min=GC_content_min, GC_content_max=GC_content_max,
Tm_min=Tm_min, Tm_max=Tm_max, min_arm_length=min_arm_length, max_arm_Tm_dif=max_arm_Tm_dif, arm_Tm_min=arm_Tm_min, arm_Tm_max=arm_Tm_max, n_jobs=4)
2023-05-26 18:41:05,992 [INFO] Parameters Property Filters:
2023-05-26 18:41:06,004 [INFO] probe_database = <oligo_designer_toolsuite.database._oligos_database.OligoDatabase object at 0x134ce32b0>
2023-05-26 18:41:06,010 [INFO] GC_content_min = 40
2023-05-26 18:41:06,012 [INFO] GC_content_max = 60
2023-05-26 18:41:06,015 [INFO] Tm_min = 52
2023-05-26 18:41:06,016 [INFO] Tm_max = 67
2023-05-26 18:41:06,017 [INFO] min_arm_length = 10
2023-05-26 18:41:06,019 [INFO] max_arm_Tm_dif = 2
2023-05-26 18:41:06,025 [INFO] arm_Tm_min = 38
2023-05-26 18:41:06,026 [INFO] arm_Tm_max = 49
2023-05-26 18:41:06,029 [INFO] Tm_parameters_probe = {'check': True, 'strict': True, 'c_seq': None, 'shift': 0, 'nn_table': 'DNA_NN3', 'tmm_table': 'DNA_TMM1', 'imm_table': 'DNA_IMM1', 'de_table': 'DNA_DE1', 'dnac1': 50, 'dnac2': 0, 'selfcomp': False, 'dNTPs': 0, 'saltcorr': 7, 'Na': 1.25, 'K': 75, 'Tris': 20, 'Mg': 10}
2023-05-26 18:41:06,030 [INFO] Tm_chem_correction_param_probe = {'DMSO': 0, 'DMSOfactor': 0.75, 'fmdfactor': 0.65, 'fmdmethod': 1, 'GC': None, 'fmd': 20}
2023-05-26 18:41:06,031 [INFO] n_jobs = 4
2023-05-26 21:18:45,849 [INFO] Step - Filter Probes by Sequence Property: the database contains 3732914 probes from 882 genes, while 32224282 probes and 5 genes have been deleted in this step.
Parameters for Specificity Filters
BlastN Similarity Filter: - blast_word_size: word size for the blastn seed (exact match to target) - blast_percent_identity: maximum similarity between oligos and target sequences, ranging from 0 to 100% (no missmatch) - blast_coverage: minimum coverage between oligos and target sequence, ranging from 0 to 100% (full coverage)
Bowtie Ligation Region filter: - ligation_region_size: size of the seed region around the ligation site for bowtie seed region filter
Note: Depending on the number of genes, this step might be time and memory consuming. For high number of genes, you might want to run this step on a bigger machine!
[9]:
####### Load existing database #######
# load annotation files for Reference Database
# source = "custom"
# custom_params = {
# "file_annotation": "./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.gtf",
# "file_sequence": "./output/annotation/GCF_000001405.40_GRCh38.p14_genomic.fna",
# "files_source": "NCBI",
# "species": "Homo_sapiens",
# "annotation_release": "110",
# "genome_assembly": "GRCh38.p14",
# }
# probe_designer.load_annotations(source=source, source_params=custom_params)
# # load existing database
# file_database = "./output/oligo_database/probe_database_property_filter.txt"
# min_probes_per_gene = 3
# probe_database = probe_designer.load_probe_database(file_database=file_database, min_probes_per_gene=min_probes_per_gene)
####### Apply Specificity Filter #######
ligation_region_size=5
blast_word_size=10
blast_percent_identity=80
blast_coverage=50
probe_database, file_database = probe_designer.filter_probes_by_specificity(probe_database, ligation_region_size=ligation_region_size,
blast_word_size=blast_word_size, blast_percent_identity=blast_percent_identity, blast_coverage=blast_coverage, n_jobs=2)
2023-05-26 21:18:46,642 [INFO] Parameters Specificity Filters:
2023-05-26 21:18:46,647 [INFO] probe_database = <oligo_designer_toolsuite.database._oligos_database.OligoDatabase object at 0x134ce32b0>
2023-05-26 21:18:46,651 [INFO] ligation_region_size = 5
2023-05-26 21:18:46,652 [INFO] blast_word_size = 10
2023-05-26 21:18:46,653 [INFO] blast_percent_identity = 80
2023-05-26 21:18:46,654 [INFO] blast_coverage = 50
2023-05-26 21:18:46,654 [INFO] n_jobs = 2
2023-05-27 01:19:33,100 [INFO] Step - Filter Probes by Specificity: the database contains 570410 probes from 796 genes, while 3162504 probes and 86 genes have been deleted in this step.
After applying different sets of filters to the probe database, we will create probesets for each gene, which are sets of probes that do not overlap and have a high efficiency score (calculated from melting temperature and GC content).
Parameters for Oligo Efficiency Score
Tm_min: minimum melting temperature of probes
Tm_max: maximum melting temperature of probes
Tm_opt: optimal melting temperature of probes
Tm_weight: weight of the Tm of the probe in the efficiency score
GC_content_min: minimum GC content of probes
GC_content_max: maximum GC content of probes
GC_content_opt: optimal GC content of probes
GC_weight: weight of the GC content of the probe in the efficiency score
Parameters for Oligosets Generation
probeset_size_opt: ideal number of oligos per probeset
probeset_size_min: minimum number of oligos per probeset
n_sets: maximum number of sets per gene
[10]:
####### Load existing database #######
# file_database = "./output/oligo_database/oligo_database_specificity_filters.txt"
# min_probes_per_gene = 3
# probe_database = probe_designer.load_probe_database(file_database=file_database, min_probes_per_gene=min_probes_per_gene)
####### Apply Probe Set Selection #######
probeset_size_opt=5
probeset_size_min=2
n_sets=100
Tm_min=52
Tm_max=67
Tm_opt=60
Tm_weight=1
GC_content_min=40
GC_content_max=60
GC_content_opt=50
GC_weight=1
probe_database, file_database, dir_oligosets = probe_designer.create_probe_sets(probe_database,
probeset_size_opt=probeset_size_opt,
probeset_size_min=probeset_size_min,
n_sets=n_sets,
Tm_min=Tm_min,
Tm_max=Tm_max,
Tm_opt=Tm_opt,
Tm_weight=Tm_weight,
GC_content_min=GC_content_min,
GC_content_max=GC_content_max,
GC_content_opt=GC_content_opt,
GC_weight=GC_weight,
n_jobs=2)
2023-05-27 01:19:33,698 [INFO] Parameters Probesets:
2023-05-27 01:19:33,702 [INFO] probe_database = <oligo_designer_toolsuite.database._oligos_database.OligoDatabase object at 0x134ce32b0>
2023-05-27 01:19:33,703 [INFO] probeset_size_opt = 5
2023-05-27 01:19:33,705 [INFO] probeset_size_min = 2
2023-05-27 01:19:33,706 [INFO] n_sets = 100
2023-05-27 01:19:33,706 [INFO] Tm_min = 52
2023-05-27 01:19:33,707 [INFO] Tm_max = 67
2023-05-27 01:19:33,708 [INFO] Tm_opt = 60
2023-05-27 01:19:33,709 [INFO] Tm_weight = 1
2023-05-27 01:19:33,710 [INFO] GC_content_min = 40
2023-05-27 01:19:33,710 [INFO] GC_content_max = 60
2023-05-27 01:19:33,711 [INFO] GC_content_opt = 50
2023-05-27 01:19:33,712 [INFO] GC_weight = 1
2023-05-27 01:19:33,713 [INFO] n_jobs = 2
2023-05-27 03:19:32,175 [INFO] Step - Generate Oligosets: the database contains 11525 probes from 756 genes, while 558885 probes and 40 genes have been deleted in this step.
[11]:
# get gene names of genes with sufficient number of probes to proceed with next step
genes_with_sufficient_probes = probe_database.database.keys()
# add extra clolumn to anndata to mark genes with sufficient probes
pbmc_data.var["sufficient_probes"] = False
for gene in highly_variable_genes:
if gene in genes_with_sufficient_probes:
pbmc_data.var["sufficient_probes"][gene] = True
# create a new variable that indicates if the gene passes the first constraint filter
pbmc_data.var["pass_constraints"] = [su_p and hi_v for su_p, hi_v in zip(pbmc_data.var["sufficient_probes"], pbmc_data.var["highly_variable"])]
ProbesetSelector class. We specify the number of genes n (20) and the keys in adata.obs and adata.var where we find the cell type annotations (celltype_key="celltype") and selected genes (celltype_key="celltype"), respectively.Executing the cell below will give us a warning that the cell type clusters for dendritic cells and megakaroycytes are quite small and therefore the genes that are selected to identify these cell types potentially don’t generalize very well. The method will not exclude these cell types automatically, but it can be done manually by setting the parameter celltypes to a subset of cell types instead of celltypes="all".
[12]:
##### Select genes for gene panel #####
selector = sp.se.ProbesetSelector(pbmc_data, n=20, genes_key="pass_constraints", celltype_key="celltype", verbosity=1, save_dir=None)
selector.select_probeset()
selected_genes = selector.probeset.index[selector.probeset.selection]
Note: The following celltypes' test set sizes for forest training are below min_test_n (=20):
Dendritic cells : 9
Megakaryocytes : 3
The genes selected for those cell types potentially don't generalize well. Find the genes for each of those cell types in self.genes_of_primary_trees after running self.select_probeset().
Once we have all selected genes, we create the final “read to order” probe sequences. Calling the fuction below will produce two files, *[padlock, merfish, seqfish]_probes* and *[padlock, merfish, seqfish]_probes_order*. The latter file contains the ready to order probe sequences for each gene.
Parameters for Padlock Final Sequence Design
detect_oligo_length_min: minimum length of detection oligo
detect_oligo_length_max: maximum length of detection oligo
detect_oligo_Tm_opt: optimal melting temperature of detection oligo
Tm_parameters_detection_oligo: melting temperature parameters for detection oligo design
Tm_chem_correction_param_detection_oligo: parameters for chemical correction of melting temperature for detection oligo design
Note: The melting temperature is used in 2 different stages (probe and detection oligo design), where a few parameters are shared and the others differ. Parameters for melting temperature -> for more information on parameters, see:here
[13]:
##### Remove all genes from the database that are not selected for the gene panel ####
probe_database.database = {key: value for key, value in probe_database.database.items() if key in selected_genes}
probe_database.oligosets = {key: value for key, value in probe_database.oligosets.items() if key in selected_genes}
[14]:
##### Design final sequences #####
detect_oligo_length_min = 18
detect_oligo_length_max = 25
detect_oligo_Tm_opt = 32
probe_designer.create_final_sequences(probe_database, detect_oligo_length_min, detect_oligo_length_max, detect_oligo_Tm_opt)
2023-05-27 03:23:40,007 [DEBUG] handle_msg[8670f070af094472be824ecf093d55a3]({'header': {'date': datetime.datetime(2023, 5, 27, 1, 23, 39, 934000, tzinfo=tzutc()), 'msg_id': '5a26b004-efe4-482a-a30b-7bed1068077b', 'msg_type': 'comm_msg', 'session': 'f80bbb6a-f083-4dbc-9c24-00aa83c84915', 'username': '16c5b64d-425a-4c50-b860-381f99d9c018', 'version': '5.2'}, 'msg_id': '5a26b004-efe4-482a-a30b-7bed1068077b', 'msg_type': 'comm_msg', 'parent_header': {}, 'metadata': {}, 'content': {'comm_id': '8670f070af094472be824ecf093d55a3', 'data': {'method': 'update', 'state': {'outputs': [{'output_type': 'display_data', 'data': {'text/plain': '\x1b[1;30mSPAPROS PROBESET SELECTION: \x1b[0m \x1b[33m0:04:06\x1b[0m\n\x1b[1;34mSelect pca genes..........................................\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m100%\x1b[0m \x1b[33m0:00:00\x1b[0m\n\x1b[1;34mTrain baseline forest based on DE genes...................\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 4/4\x1b[0m \x1b[33m0:03:12\x1b[0m\n \x1b[1;2;36mSelect DE genes.........................................\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 8/8\x1b[0m \x1b[33m0:00:00\x1b[0m\n \x1b[1;2;36mTrain prior forest for DE_baseline forest...............\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 3/3\x1b[0m \x1b[33m0:00:40\x1b[0m\n \x1b[1;2;36mIteratively add DE genes to DE_baseline forest..........\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 3/3\x1b[0m \x1b[33m0:01:41\x1b[0m\n \x1b[1;2;36mTrain final baseline forest on all celltypes............\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 3/3\x1b[0m \x1b[33m0:00:49\x1b[0m\n\x1b[1;34mTrain final forests.......................................\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 3/3\x1b[0m \x1b[33m0:00:53\x1b[0m\n \x1b[1;2;36mTrain forest on pre/prior/pca selected genes............\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m 3/3\x1b[0m \x1b[33m0:00:53\x1b[0m\n \x1b[1;2;36mInitial results are good enough. No genes are added.......................................\x1b[0m \n\x1b[1;34mCompile probeset list.....................................\x1b[0m \x1b[38;2;114;156;31m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\x1b[0m \x1b[35m100%\x1b[0m \x1b[33m0:00:00\x1b[0m\n\x1b[1;30mFINISHED\x1b[0m \n \n', 'text/html': '<pre style="white-space:pre;overflow-x:auto;line-height:normal;font-family:Menlo,\'DejaVu Sans Mono\',consolas,\'Courier New\',monospace"><span style="color: #000000; text-decoration-color: #000000; font-weight: bold">SPAPROS PROBESET SELECTION: </span> <span style="color: #808000; text-decoration-color: #808000">0:04:06</span>\n<span style="color: #000080; text-decoration-color: #000080; font-weight: bold">Select pca genes..........................................</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080">100%</span> <span style="color: #808000; text-decoration-color: #808000">0:00:00</span>\n<span style="color: #000080; text-decoration-color: #000080; font-weight: bold">Train baseline forest based on DE genes...................</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 4/4</span> <span style="color: #808000; text-decoration-color: #808000">0:03:12</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Select DE genes.........................................</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 8/8</span> <span style="color: #808000; text-decoration-color: #808000">0:00:00</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Train prior forest for DE_baseline forest...............</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 3/3</span> <span style="color: #808000; text-decoration-color: #808000">0:00:40</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Iteratively add DE genes to DE_baseline forest..........</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 3/3</span> <span style="color: #808000; text-decoration-color: #808000">0:01:41</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Train final baseline forest on all celltypes............</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 3/3</span> <span style="color: #808000; text-decoration-color: #808000">0:00:49</span>\n<span style="color: #000080; text-decoration-color: #000080; font-weight: bold">Train final forests.......................................</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 3/3</span> <span style="color: #808000; text-decoration-color: #808000">0:00:53</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Train forest on pre/prior/pca selected genes............</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080"> 3/3</span> <span style="color: #808000; text-decoration-color: #808000">0:00:53</span>\n <span style="color: #7fbfbf; text-decoration-color: #7fbfbf; font-weight: bold">Initial results are good enough. No genes are added.......................................</span> \n<span style="color: #000080; text-decoration-color: #000080; font-weight: bold">Compile probeset list.....................................</span> <span style="color: #729c1f; text-decoration-color: #729c1f">━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━</span> <span style="color: #800080; text-decoration-color: #800080">100%</span> <span style="color: #808000; text-decoration-color: #808000">0:00:00</span>\n<span style="color: #000000; text-decoration-color: #000000; font-weight: bold">FINISHED</span> \n \n</pre>\n'}, 'metadata': {}}]}, 'buffer_paths': []}}, 'buffers': []})
2023-05-27 03:23:40,069 [INFO] Parameters Final Sequence Design:
2023-05-27 03:23:40,070 [INFO] probe_database = <oligo_designer_toolsuite.database._oligos_database.OligoDatabase object at 0x134ce32b0>
2023-05-27 03:23:40,072 [INFO] detect_oligo_length_min = 18
2023-05-27 03:23:40,073 [INFO] detect_oligo_length_max = 25
2023-05-27 03:23:40,074 [INFO] detect_oligo_Tm_opt = 32
2023-05-27 03:23:40,076 [INFO] Tm_parameters_detection_oligo = {'check': True, 'strict': True, 'c_seq': None, 'shift': 0, 'nn_table': 'DNA_NN3', 'tmm_table': 'DNA_TMM1', 'imm_table': 'DNA_IMM1', 'de_table': 'DNA_DE1', 'dnac1': 50, 'dnac2': 0, 'selfcomp': False, 'dNTPs': 0, 'saltcorr': 7, 'Na': 39, 'K': 0, 'Tris': 0, 'Mg': 0}
2023-05-27 03:23:40,077 [INFO] Tm_chem_correction_param_detection_oligo = {'DMSO': 0, 'DMSOfactor': 0.75, 'fmdfactor': 0.65, 'fmdmethod': 1, 'GC': None, 'fmd': 30}
2023-05-27 03:23:40,347 [INFO] Step - Design Final Padlock Sequences: padlock sequences are stored in './output/padlock_sequences/padlock_sequences' directory.